KAIST
BREAKTHROUGHS
Research Webzine of the KAIST College of Engineering since 2014
Fall 2024 Vol. 23Computational prediction of metabolites associated with somatic mutations in cancers
KAIST researchers developed a computational workflow to predict cancer-related metabolites and pathways using patient-specific metabolic models and somatic mutation data, aiding the identification of novel oncometabolites and advancing cancer treatment strategies
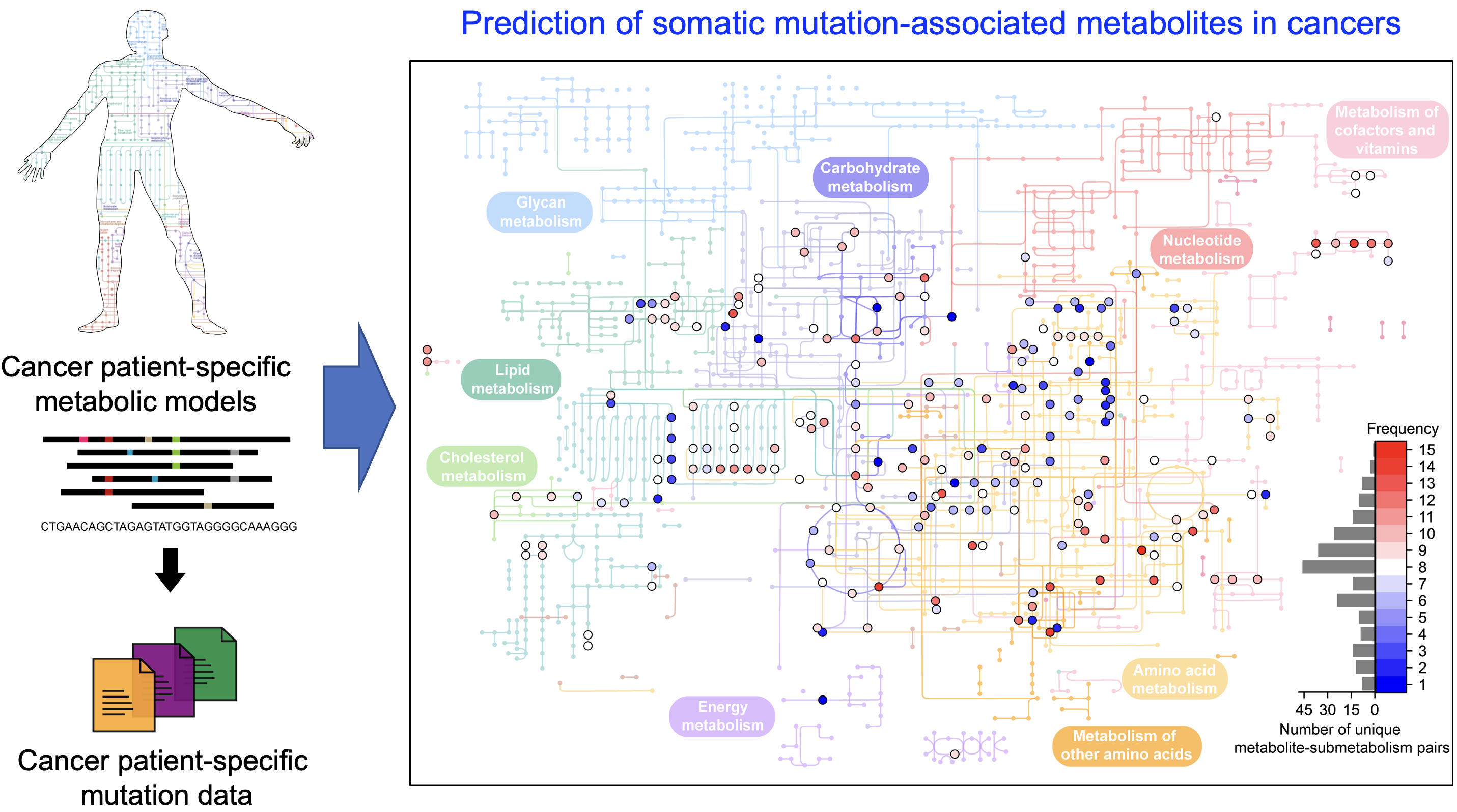
Cancer is characterized by abnormal metabolic processes that differ significantly from those of normal cells. Consequently, extensive research has been devoted to understanding cancer metabolism to develop effective diagnostic and therapeutic strategies. Noteworthy achievements in this field include the discovery of oncometabolites—metabolites that exhibit pro-oncogenic functions when abnormally accumulated in cancer cells. Oncometabolites are often produced as a result of gene mutations, promoting the growth and survival of cancer cells. Examples of oncometabolites include 2-hydroxyglutarate, succinate, and fumarate. The U.S. Food and Drug Administration (FDA) has approved anticancer drugs such as Tibsovo (active ingredient: ivosidenib) and Idhifa (active ingredient: enasidenib), both used to treat acute myeloid leukemia by targeting enzymes associated with oncometabolites.
Despite these advancements, studying cancer metabolism, particularly oncometabolites, remains challenging due to the time-consuming and costly nature of methodologies such as metabolomics. As a result, although many cancer-associated gene mutations have been extensively studied, there remain few confirmed oncometabolites.
A research team from KAIST led by Professor Hyun Uk Kim in the Department of Chemical and Biomolecular Engineering developed a computational workflow that systematically predicts metabolites and metabolic pathways associated with somatic mutations in cancer. This breakthrough was achieved in collaboration with research teams led by Professors Youngil Koh, Hongseok Yun, and Chang Wook Jeong from Seoul National University Hospital.
The research teams successfully reconstructed patient-specific genome-scale metabolic models (GEMs) for 1,043 cancer patients across 24 cancer types. GEMs are computational models that mathematically describe all of the biochemical reactions occurring within a cell, allowing for the prediction of the cell’s metabolic phenotypes under various genetic and environmental conditions. The teams integrated publicly available transcriptome data from international cancer genome consortiums such as PCAWG and TCGA into a generic human GEM, making it possible to predict each patient’s metabolic phenotypes.
Using these patient-specific GEMs, the team developed a four-step computational workflow. This workflow begins with the calculation of the flux-sum value of each metabolite by simulating the patient-specific GEMs. The flux-sum value quantifies the intracellular importance of a metabolite. The next step involves statistically analyzing the predicted flux-sum data alongside mutation data to identify metabolites significantly associated with specific gene mutations. Finally, the workflow selects altered metabolic pathways that significantly contribute to the biosynthesis of the predicted oncometabolite candidates, producing metabolite-gene-pathway sets as the output.
The computational workflow thus developed can systematically predict how genetic mutations impact cellular metabolism through metabolic pathways. Importantly, it is readily applicable to different cancer types based on mutation and transcriptome data from cancer patient cohorts. The computational workflow and its predictive outcomes will serve as the groundwork for identifying novel oncometabolites and facilitating the development of various treatment and diagnostic strategies.
This study has been published in Genome Biology, a representative journal in the field of biotechnology and genetics (https://genomebiology.biomedcentral.com/articles/10.1186/s13059-024-03208-8).
Most Popular

High-performance and sustainable paper coating material that prevents microplastics
Read more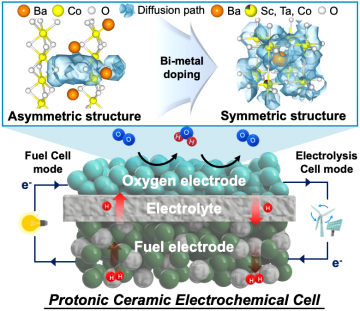
Next-generation ceramic electrochemical cells for global net-zero goals
Read more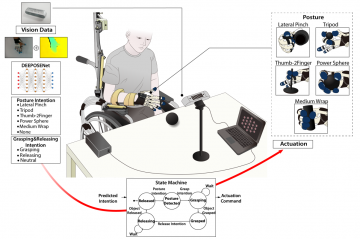
Multi-hand posture rehabilitation for stroke survivors: Rehabilitation system using vision-based intention recognition and a soft robotic glove
Read more
Impacts of new town developments on carbon sinks in the Seoul metropolitan area
Read more
Revolutionizing strength: Hercules artificial muscles 17 times stronger than human muscles
Read more