KAIST
BREAKTHROUGHS
Research Webzine of the KAIST College of Engineering since 2014
Fall 2025 Vol. 25Structural insight into the molecular mechanism of PET degradation
Structural insight into the molecular mechanism of PET degradation
A superior poly ethylene terephthalate (PET)-degrading enzyme was developed using structural-based protein engineering and suggests a molecular mechanism for superior degradability of PET. This study will lead to developing further superior enzymes and constructing platforms for microbial plastic recycling.
Article | Fall 2019
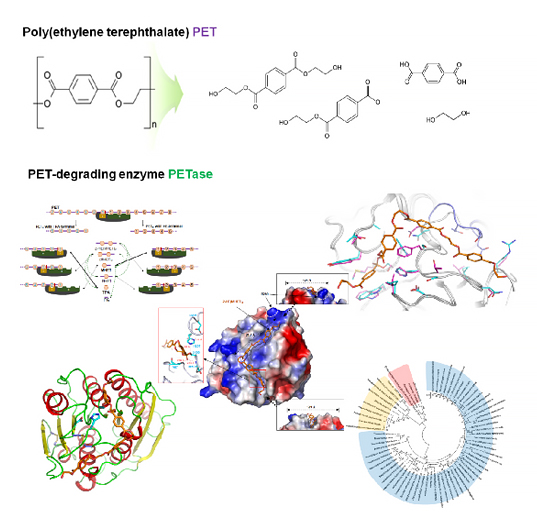
A KAIST metabolic engineering research team has newly suggested a molecular mechanism showing superior degradability of poly ethylene terephthalate (PET). This is the first report to simultaneously determine the 3D crystal structure of Ideonella sakaiensis PETase and develop a new variant with enhanced PET degradation.
Recently, diverse research projects are working to address the non-degradability of materials. A poly ethylene terephthalate (PET)-degrading bacterium called Ideonella sakaiensis was recently identified for the possible degradation and recycling of PET by a Japanese team whose work was published in the journal Science (Yoshida et al., 2016). The PETase of I. sakaiensis (IsPETase) can degrade PET more successfully in the ambient temperature than other PET-degrading enzymes. However, the detailed molecular mechanism of PET degradation had not been elucidated, hindering further studies.
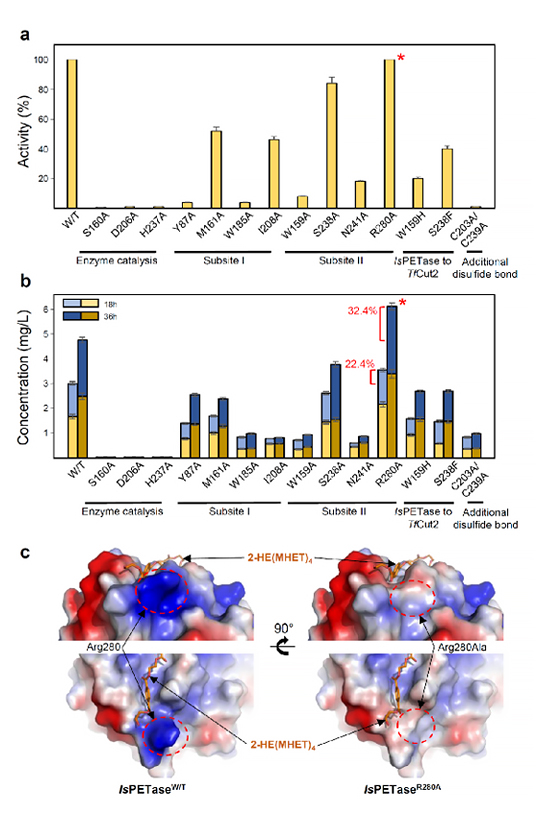
A team under Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering and a team under Professor Kyung-Jin Kim of the Department of Biotechnology at Kyungpook National University investigated how the substrate binds to the enzyme and which differences in enzyme structure result in significantly higher PET degrading activity compared with other cutinases and esterases, which make IsPETase highly attractive for industrial applications toward PET waste recycling. Based on the 3D structure and related biochemical studies, they successfully predicted the reasons for extraordinary PET degrading activity of IsPETase and suggested other enzymes that can degrade PET with a newly classified phylogenetic tree.
The research teams predicted a special molecular mechanism based on the docking simulation between PETase and a PET alternative mimic substrate. The team proposed that 4 MHET moieties are the most properly matched substrates due to a cleft on structure even with the 10-20-mers for PET. This is meaningful in that it is the first docking simulation between PETase and PET, not its monomer.
Furthermore, they succeeded in constructing a variant for IsPETase with enhanced PET-degrading activity using structural-based protein engineering. They also determined a crystal structure of this variant to show that the changed structure is better to accommodate PET substrates than wild-type PETase, which will lead to developing further superior enzymes and constructing platforms for microbial plastic recycling.
It is expected that the new approaches taken in this research can be background for further study of other enzymes capable of degrading not only PET but other plastics as well.
Professor Lee said, “Environmental pollution from plastics remains one of the greatest challenges worldwide with the increasing consumption of plastics. We successfully constructed a new superior PET-degrading variant with the determination of a crystal structure of PETase and its degrading molecular mechanism. This novel technology will help further studies to engineer superior enzymes with high efficiency in degrading. This will be the subject of our team’s ongoing research projects to address the global environmental pollution problem for next generation.”
This research was published in Nature Comm., 9:382, DOI: 10.1038/s41467-018-02881-1, and was also highlighted by a number of press media.
Further Contact: Dr. Sang Yup Lee, Distinguished Professor, KAIST, Daejeon, Korea (leesy@kaist.ac.kr, +82-42-350-3930)
Journal reference
Joo, S., Cho, I.J., Seo, H., Son, H.F., Sagong, H.-Y., Shin, T.J., Choi, S.Y., Lee, S.Y. & Kim, K.J., “Structural insight into molecular mechanism of poly (ethylene terephthalate) degradation”, Nature Comm., 9:382, DOI: 10.1038/s41467-018-02881-1 (2018.1)
Most Popular

A New solution enabling soft growing robots to perform a variety of tasks in confined spaces
Read more
Towards a more reliable evaluation system than humans - BiGGen-Bench
Read more
Development of a compact high-resolution spectrometer using a double-layer disordered metasurface
Read more
AI-Designed carbon nanolattice: Feather-light, steel-strong
Read more
Dual‑Mode neuransistor for on‑chip liquid‑state computing
Read more