KAIST
BREAKTHROUGHS
Research Webzine of the KAIST College of Engineering since 2014
Spring 2025 Vol. 24AI now assists pathologists in detecting cancerous cells and making accurate diagnoses.
AI now assists pathologists in detecting cancerous cells and making accurate diagnoses.
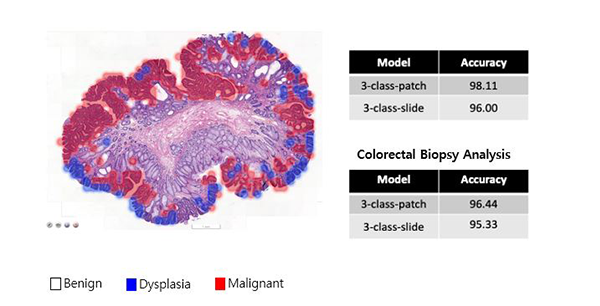
AI diagnosis models achieve accuracy rates of 96.0% and 95.3% when used to classify histopathological images of gastric and colorectal endoscopic biopsy specimens, respectively, into three classes. They are currently in service, assisting pathologists in the field, as the core components of a post-analytic daily quality-control system.
Article | Fall 2022
Despite the rapid advances in artificial intelligence (AI), it is still challenging to automatically diagnose heterogeneous diseases that can lead to loss of life. Cancer is one such highly heterogeneous disease and one of the leading causes of death, ranking second in deaths per year globally [1]. To diagnose the presence of cancer, pathologists usually examine whole-slide images (WSIs) to identify abnormal cells. The growth in the number of yearly cancer cases has led to expert pathologists working long hours, thereby increasing the chance of human errors, which reportedly ranges from 3% to 9% in anatomical pathology [2]. To alleviate this problem, AI-based, in particular deep-learning-based, diagnosis systems for WSI analysis have been developed to assist pathologists in the field.
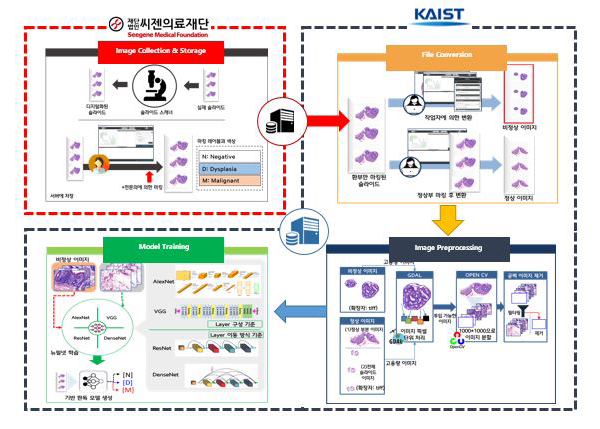
Researchers at the Knowledge Innovation Research Center (KIRC) at KAIST have been working with the Seegene Medical Foundation, one of the largest diagnosis and pathology institutions in South Korea, to develop an AI-based cancer detection and diagnosis system over the past four years. As shown in Figure 1, in this collaborative project, Seegene is responsible for image collection, image labelling and annotation, and storage management while KIRC@KAIST is responsible for the subsequent processing of the images for AI model development.
A whole-slide image (WSI) of biopsy tissue consists of many gigapixels of data (typically 50,00050,000 pixels). Given such a large size, it is difficult to process a WSI directly. Thus, we have designed a novel patch-based framework in which a WSI is split into patches, the latent features from the patches are extracted for a patch-level classification model, and the patch-level features are aggregated for a slide-level classification model. While primarily leveraging the CNN (convolutional neural network) technique, the framework pays special attention to issues unique to the pathology domain, such as the differential severity of Type I vs Type II errors, the locality of pixel dependencies, and unspecialized orientations of slide images.
In a lab test of the developed models using balanced datasets, the models achieved accuracy rates of 96.0% and 95.3% in classifying histopathological WSIs of gastric and colorectal endoscopic biopsy specimens, respectively, into three classes (negative, dysplasia, malignant). The models are currently in service, assisting pathologists in the field, as the core components of a post-analytic daily quality-control system, which means that the pathologists’ decisions are crossed-checked by the system to detect any potential human errors early. The system not only determines the presence of cancer but also identifies problematic areas, as shown in Figure 2.
The collaborative project has been found mutually fruitful. Researchers at Seegene and KIRC@KAIST have been working in unison to make academic and practical contributions, with a number of tangible outcomes, including a paper recently published in Scientific Reports [3] and a domestic patent registered [4]. They have also jointly filed multiple international patents, which are currently under review. During the three-month AI-assisted daily quality-control trial run period, approximately 17–30 times the number of current potential human errors could be detected within an average of 1.2 days. Consequently, active measures have been made possible before pathology report errors are relayed to the patients.
References
1. Siegel, R.L., K.D. Miller, H.E. Fuchs, and A. Jemal, Cancer statistics, 2021. CA: A cancer journal for clinicians, 2021. 71(1): p. 7-33.
2. Peck, M., D. Moffat, B. Latham, and T. Badrick, Review of diagnostic error in anatomical pathology and the role and value of second opinions in error prevention. Journal of clinical pathology, 2018. 71(11): p. 995-1000.
3. Ashraf, M., W.R.Q. Robles, M. Kim, Y.S. Ko, and M.Y. Yi, A loss-based patch label denoising method for improving whole-slide image analysis using a convolutional neural network, Scientific Reports, 2022-01.
4. Yi, M.Y., Y.J. Park, J.K. Chun, and Y.S. Ko, Deep Learning Model Based Image Diagnosis Apparatus and Method Thereof (Korea Patent No. 10-2132375-0000), Korea Intellectual Property Office, 2020-07-03.
Most Popular

When and why do graph neural networks become powerful?
Read more
Smart Warnings: LLM-enabled personalized driver assistance
Read more
Extending the lifespan of next-generation lithium metal batteries with water
Read more
Professor Ki-Uk Kyung’s research team develops soft shape-morphing actuator capable of rapid 3D transformations
Read more
Oxynizer: Non-electric oxygen generator for developing countries
Read more