KAIST
BREAKTHROUGHS
Research Webzine of the KAIST College of Engineering since 2014
Spring 2025 Vol. 24Bio-integrated microfluidics chips: Rapid investigation of combinatory antibiotic effects against pathogenic bacteria
Bio-integrated microfluidics chips: Rapid investigation of combinatory antibiotic effects against pathogenic bacteria
A microfluidic chip significantly reduces the turnaround time and labor-intensive work of combinatory antibiotic screening against bacteria by generating two orthogonal concentration gradients in a diffusion manner.
Article | Fall 2019
Over the past few decades, antibiotic-resistant bacteria have been identified as a significant challenge. In 2014, the World Health Organization (WHO) officially reported that the antibiotic resistance of bacteria had reached to a level of serious threat. Combinatory therapy, which combines two or more antibiotics, has been presented as a means to overcome pathogenic bacteria. Although the therapy mostly showed enhanced antimicrobial effects, some pairs resulted in unfavorable outcomes such as suppressed bactericidal activities. Therefore, combinatory screening is a crucial preliminary process to find suitable antibiotic pairs and their concentration range against unknown pathogens. However, the conventional platforms are inconvenient for concentration dilution and sample preparation, and they take more than 24 hours to produce results.
A KAIST research team lead by Jessie Sungyun Jeon, an assistant professor of the Department of Mechanical Engineering, collaborated with Professor Hyun Jung Chung from the Department of Biological Sciences to develop a microfluidic chip that identifies pharmacological interactions of an antibiotic pair to address the aforementioned issues.
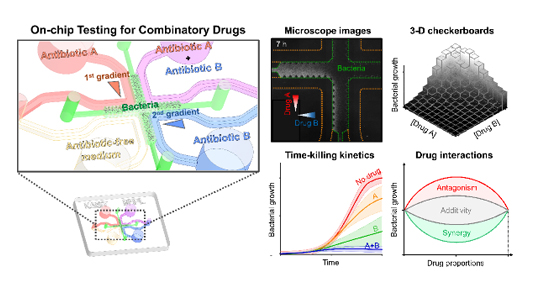
To investigate the combinatory effects, the team loaded a mixture of bacterial samples and agarose into the central microchannel and injected reagents with or without antibiotics into four surrounding microchannels (Figure 1). The diffusion of antibiotic molecules from the channel with antibiotics to the channel without antibiotics resulted in the formation of two orthogonal concentration gradients of the two antibiotics on the bacteria-trapped agarose.
The team observed the inhibition of bacterial growth by the two antibiotic gradients with a microscope and analyzed the spatiotemporal changes of bacterial growth measured by a grayscale of the images (Figure 1). They confirmed different pharmacological patterns depending on the antibiotic pairs and classified the pairs into either synergy or antagonism.
Compared to the conventional platforms (e.g. 96-well plates), the chip exhibited a number of advantages which include requiring only a small amount of bacteria (~10 microliters), the instantaneous formation of 121 pairwise drug concentrations (~35 minutes), and rapid identification of drug efficacy and interactions (~8 hours).
The research team expects that the microfluidic chips could be utilized as alternatives to the existing macro-scale screening methods with potential applications in the guidance of clinical therapies and in screening other cell-type agents.
This research was published in Lab on a Chip as a back cover article (S. Kim et al, Lab Chip, 19(6), 959-973, 2019).
Most Popular

When and why do graph neural networks become powerful?
Read more
Extending the lifespan of next-generation lithium metal batteries with water
Read more
Professor Ki-Uk Kyung’s research team develops soft shape-morphing actuator capable of rapid 3D transformations
Read more
Smart Warnings: LLM-enabled personalized driver assistance
Read more
Development of a nanoparticle supercrystal fabrication method using linker-mediated covalent bonding reactions
Read more