KAIST
BREAKTHROUGHS
Research Webzine of the KAIST College of Engineering since 2014
Spring 2025 Vol. 24
A joint research team has developed DeepECtransformer, an AI that can predict enzyme functions from protein sequences. The joint research team discovered previously unknown enzymes using the AI.
Enzymes are proteins that catalyze biological reactions. Identifying the function of each enzyme is essential to understanding the various chemical reactions that exist in living organisms and the metabolic characteristics of those organisms. While Escherichia coli is one of the most studied organisms, the function of 30% of the proteins that make up E. coli has not yet been revealed. For this, a newly developed artificial intelligence (AI) model was used to discover 464 enzymes from the unknown proteins.
A joint research team comprised of Gi Bae Kim, Ji Yeon Kim, Dr. Jong An Lee and Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering at KAIST, and Dr. Charles J. Norsigian and Professor Bernhard O. Palsson of the Department of Bioengineering at UCSD have developed DeepECtransformer, an AI that can predict the enzyme functions from the protein sequence. The team has established a prediction system by utilizing the AI to quickly and accurately identify the Enzyme Commission (EC) number. EC number is an enzyme function classification system designed by the International Union of Biochemistry and Molecular Biology, and to understand the metabolic characteristics of various organisms, it is necessary to develop a technology that can quickly analyze enzymes and EC numbers of the enzymes present in the genome. Various methodologies based on deep learning have been developed to analyze the features of biological sequences, including protein function prediction. However, most have a problem of a black box, where the inference process of AI cannot be interpreted. Various prediction systems that utilize AI for enzyme function prediction have also been reported but they do not solve the black box problem, nor can they interpret the reasoning process at a fine-grained level (e.g., the level of amino acid residues in the enzyme sequence).
The joint team developed DeepECtransformer, an AI that utilizes deep learning and a protein homology analysis module, to predict the enzyme function of a given protein sequence. To better understand the features of protein sequences, the transformer architecture, which is commonly used in natural language processing, was additionally used. This was done to extract important features about enzyme functions in the context of the entire protein sequence, which enabled the team to accurately predict the EC number of the enzyme. The developed DeepECtransformer can predict a total of 5,360 EC numbers. By utilizing the prediction system, the joint research team predicted 464 enzymes of E. coli that had not yet been identified.
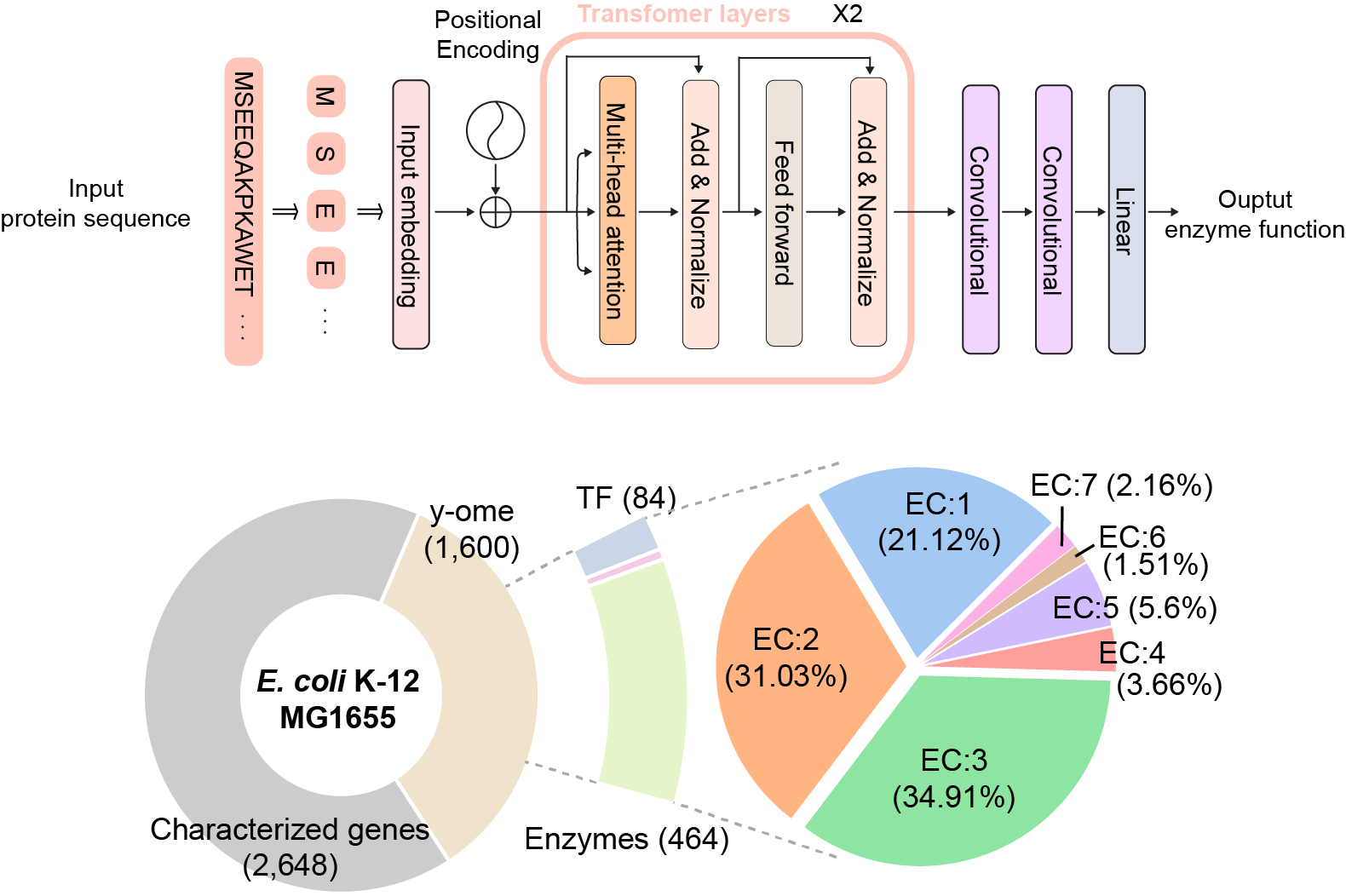
The joint team further analyzed the transformer architecture to understand the inference process of DeepECtransformer, and found that in the inference process, the AI utilizes information on catalytic active sites and/or the cofactor binding sites, which are important for enzyme function. By analyzing the black box of DeepECtransformer, it was confirmed that the AI was able to identify the features that are important for enzyme function on its own during the learning process.
Most Popular

When and why do graph neural networks become powerful?
Read more
Extending the lifespan of next-generation lithium metal batteries with water
Read more
Professor Ki-Uk Kyung’s research team develops soft shape-morphing actuator capable of rapid 3D transformations
Read more
Smart Warnings: LLM-enabled personalized driver assistance
Read more
Development of a nanoparticle supercrystal fabrication method using linker-mediated covalent bonding reactions
Read more